 2021-02-22 20:39:32
2021-02-22 20:39:32
xDNN for SARS-CoV-2 identification in patient CT scans
This article is based on the work of Nitin Mane and his GitHub release - SARS-CoV-2 xDNN Classifier. The original dataset (1) was published from the Lancaster university and it is accessible here.
The dataset and the original model were collected and developed by Asociación de Investigacion en Inteligencia Artificial Para la Leucemia Peter Moss collaborators, PlamenLancaster: Professor Plamen Angelov from Lancaster University/ Centre Director @ Lancaster Intelligent, Robotic and Autonomous systems (LIRA) Research Centre, & his researcher, Eduardo Soares PhD.
After December 2019 there was a Covid Outbreak (SARS-CoV-2), the World Health Organization (WHO) declared in the following January a global emergency. The science community replied fast and steadily to this new threat. Many datasets and information were quickly made available with an unseen pace.
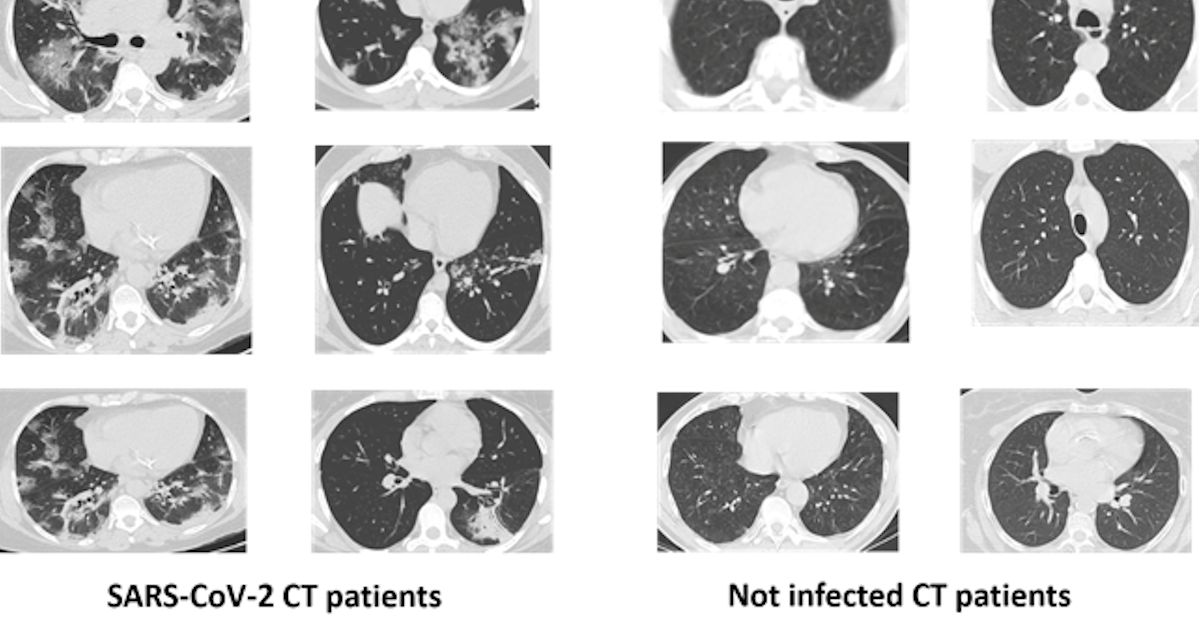
SARS-CoV-2 is inducing extensive lung damage in a relevant percentage of patients. This is often leading in respiratory impairment and successive intensive care unit (ICU) treatment. Computed tomography (CT) revealed opacity on 98% of chest CTs in infected patients. Thoracic radiology and other medical image techniques are key in evaluate the disease stage of the patient. A quick diagnosis is fundamental to start a prompt treatment that increase chances of survival.
The dataset is composed of 2482 CT scans images, which are divided in 1230 CT images from not infected patients (but they have diagnosed with other pulmonary diseases) and 1252 from SARS-CoV-2 infected patients. The dataset was collected in Brazil (hospitals from Sao Paulo). In the dataset are not listed the detailed clinical characteristics of the patients, due to ethical motivation. The dataset is composed by 60 SARS-CoV-2 infected patients and 60 not-infected patients. Overall, 32 male and 28 female SARS-CoV-2 patients where enrolled in the study, while there are 30 male and 30 female not-infected patients.
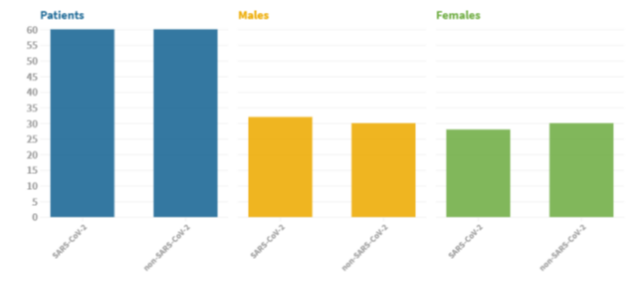
Fig2. Dataset Patient characteristic. Image from(1)
In the original article, the proposed model is trained using the images in the dataset. They used transfer learning to extract features (they used an already trained model as a feature extractor). In this case they used the VGG16 model. VGG16 is a convolutional neural network proposed by researchers in Oxford and trained on ImageNet (a large dataset of more than 14 million images and about 1000 categories). The VGG16 model achieved 92.7 % accuracy (top5) in the 2014 competition. In comparison with the previous state of the art models, VGG16 was very depth with very small convolution filters (3x3)(2). VGG16 model is widely used and it is present with the pre-trained weights in the principal deep learning library (keras, Pytorch and so on).
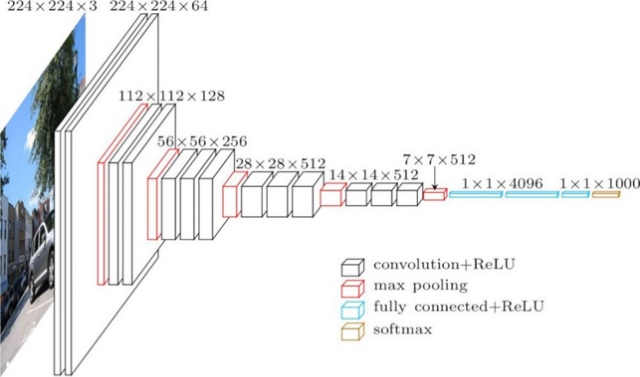
Fig 3. VGG16 architecture. Figure source: (3)
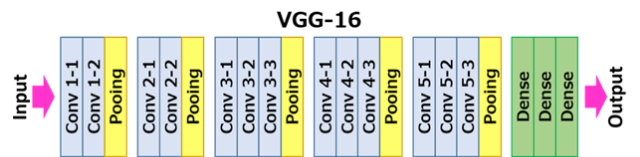
Fig 4. VGG16 scheme. (3)
The input size for the first layer (Conv 1–1) is 224 x 224 RGB images (224, 224, 3) and padding of 1. The first passage of the filters (3x3) is capable to capture spatial notions as left/right, up/down, center. Pooling is conducted as Max pooling (2x2 windows) and with stride 2. After the convolutional and pooling layers there three fully connected layers (indicated in the scheme as Dense). 4096 units for the first two layer and the last is 1000 units for the last (1 for each of the category in ImageNet). The final layer is a softmax layer.
In the original paper, they used the first dense layer to obtain feature vectors for the dataset images.
The extracted vectors are then clustered.
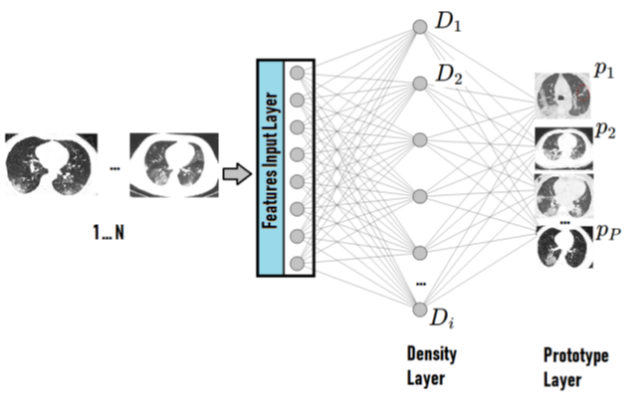
The original model has been done in Matlab an there is also a guide done on Matlab on github done by Nitin Mane and Aniruddh Sharma and can be found here
I will focus on the Python version.
To install the classifier, you need as requisite to have installed:
In the Anaconda IDE navigate to the command prompt. Anaconda use as default Python 3.7, but the project has been designed in Python 3.6 , so you need to create a new environment for Python 3.6. Then you can download the required dependencies.
conda create -n myenv python=3.6
pip -r requirements.txt
For the data pre-processing run Feature_Extraction_VGG16.py(Feature_Extraction_VGG16.py) or Feature_Extraction_VGG16_PyTorch.py(Feature_Extraction_VGG16_PyTorch.py) file in spyder.
The images are pre-processed and resized to 224 x 224 since VGG16 requires as input images of that size. Using scikit-learn the dataset is split in train and test set (20 % of the total). The VGG16 is model is therefore extracting the feature vector.
The extracted features will be shown like this
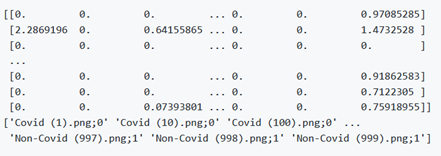
The main.py file is for the xDNN training
The xDNN is based on hierarchical clustering of Mikowski metric which process data peaks in the 2-dimension space and the key roles are the image features and labels extracted by the model.
As stated in the original article: “The proposed approach, xDNN is using prototypes. Prototypes are actual training data samples (images), which are local peaks of the empirical data distribution called typicality as well as of the data density. This generative model is identified in a closed form and equates to the pdf but is derived automatically and entirely from the training data with no user- or problem-specific thresholds, parameters or intervention. The proposed xDNN offers a new deep learning architecture that combines reasoning and learning in a synergy. It is non-iterative and non-parametric, which explains its efficiency in terms of time and computational resources. From the user perspective, the proposed approach is clearly understandable to human users.”
The results are showed in the following figures.
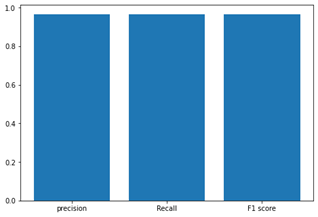
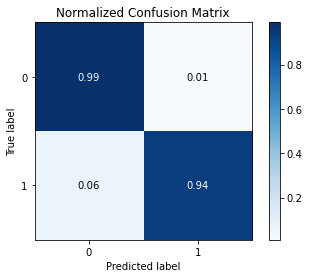
Fig6. Accuracy, Precision, Recall and F1 and Confusion Matrix. Source the GitHub repository.
Bibliography
1. Soares E, Angelov P, Biaso S, Froes MH, Abe DK. SARS-CoV-2 CT-scan dataset: A large dataset of real patients CT scans for SARS-CoV-2 identification. medRxiv (2020)2020.04.24.20078584. doi:10.1101/2020.04.24.20078584
2. Simonyan K, Zisserman A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv:14091556 [cs] (2015) Available at: http://arxiv.org/abs/1409.1556 [Accessed February 10, 2021]
3. VGG16 — Convolutional Network for Classification and Detection. (2018) Available at: https://neurohive.io/en/popular-networks/vgg16/ [Accessed February 10, 2021]